Abstract
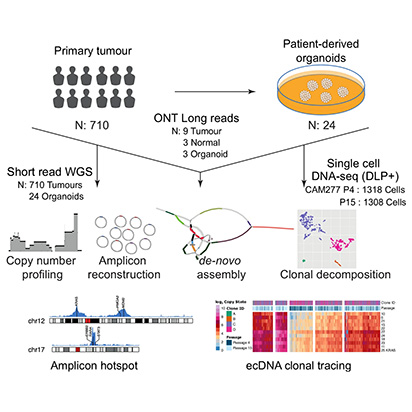
Esophageal adenocarcinoma is a prominent example of cancer characterized by frequent amplifications in oncogenes. However, the mechanisms leading to amplicons that involve breakage-fusion-bridge cycles and extrachromosomal DNA are poorly understood. Here, we use 710 esophageal adenocarcinoma cases with matched samples and patient-derived organoids to disentangle complex amplicons and their associated mechanisms. Short-read sequencing identifies ERBB2, MYC, MDM2, and HMGA2 as the most frequent oncogenes amplified in extrachromosomal DNAs. We resolve complex extrachromosomal DNA and breakage-fusion-bridge cycles amplicons by integrating of de-novo assemblies and DNA methylation in nine long-read sequenced cases. Complex amplicons shared between precancerous biopsy and late-stage tumor, an enrichment of putative enhancer elements and mobile element insertions are potential drivers of complex amplicons’ origin. We find that patient-derived organoids recapitulate extrachromosomal DNA observed in the primary tumors and single-cell DNA sequencing capture extrachromosomal DNA-driven clonal dynamics across passages. Prospectively, long-read and single-cell DNA sequencing technologies can lead to better prediction of clonal evolution in esophageal adenocarcinoma.