Abstract
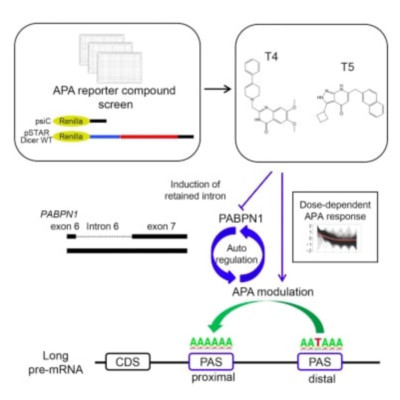
Alternative polyadenylation (APA) plays a critical role in regulating gene expression. However, the balance between genome-encoded APA processing and autoregulation by APA modulating RNA binding protein (RBP) factors is not well understood. We discovered two potent small-molecule modulators of APA (T4 and T5) that promote distal-to-proximal (DtoP) APA usage in multiple transcripts. Monotonically responsive APA events, induced by short exposure to T4 or T5, were defined in the transcriptome, allowing clear isolation of the genomic sequence features and RBP motifs associated with DtoP regulation. We found that longer vulnerable introns, enriched with distinctive A-rich motifs, were preferentially affected by DtoP APA, thus defining a core set of genes with genomically encoded DtoP regulation. Through APA response pattern and compound-small interfering RNA epistasis analysis of APA-associated RBP factors, we further demonstrated that DtoP APA usage is partly modulated by altered autoregulation of polyadenylate binding nuclear protein-1 signaling.