Abstract
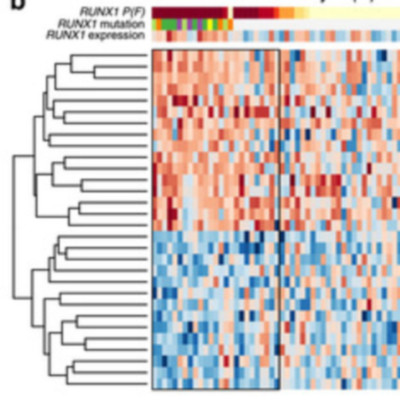
We present a novel hierarchical Bayes statistical model, xseq, to systematically quantify the impact of somatic mutations on expression profiles. We establish the theoretical framework and robust inference characteristics of the method using computational benchmarking. We then use xseq to analyse thousands of tumour data sets available through The Cancer Genome Atlas, to systematically quantify somatic mutations impacting expression profiles. We identify 30 novel cis-effect tumour suppressor gene candidates, enriched in loss-of-function mutations and biallelic inactivation. Analysis of trans-effects of mutations and copy number alterations with xseq identifies mutations in 150 genes impacting expression networks, with 89 novel predictions. We reveal two important novel characteristics of mutation impact on expression: (1) patients harbouring known driver mutations exhibit different downstream gene expression consequences; (2) expression patterns for some mutations are stable across tumour types. These results have critical implications for identification and interpretation of mutations with consequent impact on transcription in cancer.