Abstract
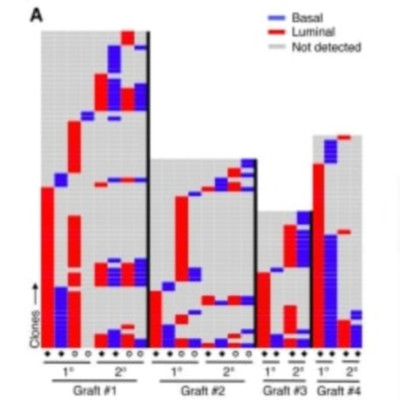
Cellular barcoding offers a powerful approach to characterize the growth and differentiation activity of large numbers of cotransplanted stem cells. Here, we describe a lentiviral genomic-barcoding and analysis strategy and its use to compare the clonal outputs of transplants of purified mouse and human basal mammary epithelial cells. We found that both sources of transplanted cells produced many bilineage mammary epithelial clones in primary recipients, although primary clones containing only one detectable mammary lineage were also common. Interestingly, regardless of the species of origin, many clones evident in secondary recipients were not detected in the primary hosts, and others that were changed from appearing luminal-restricted to appearing bilineage. This barcoding methodology has thus revealed conservation between mice and humans of a previously unknown diversity in the growth and differentiation activities of their basal mammary epithelial cells stimulated to grow in transplanted hosts.