Abstract
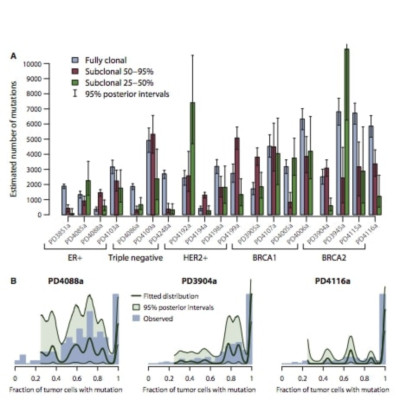
Cancer evolves dynamically as clonal expansions supersede one another driven by shifting selective pressures, mutational processes, and disrupted cancer genes. These processes mark the genome, such that a cancer’s life history is encrypted in the somatic mutations present. We developed algorithms to decipher this narrative and applied them to 21 breast cancers. Mutational processes evolve across a cancer’s lifespan, with many emerging late but contributing extensive genetic variation. Subclonal diversification is prominent, and most mutations are found in just a fraction of tumor cells. Every tumor has a dominant subclonal lineage, representing more than 50% of tumor cells. Minimal expansion of these subclones occurs until many hundreds to thousands of mutations have accumulated, implying the existence of long-lived, quiescent cell lineages capable of substantial proliferation upon acquisition of enabling genomic changes. Expansion of the dominant subclone to an appreciable mass may therefore represent the final rate-limiting step in a breast cancer’s development, triggering diagnosis.